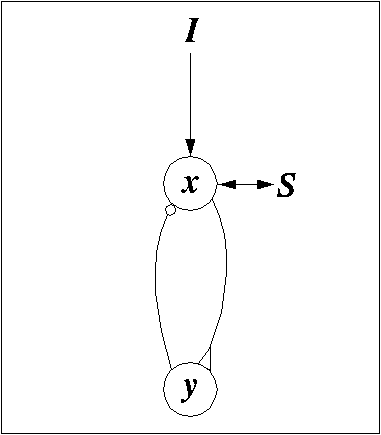
FIGURE 1. Diagram of a single oscillator with an excitatory unit x and an inhibitory unit y in a feedback loop. A triangle indicates an excitatory connection and a circle indicates an inhibitory connection. I indicates external input and S indicates the coupling with the rest of the network.

FIGURE 2. A 2D LEGION network with 4-neighborhood connections. The global inhibitor is indicated with a black circle and is coupled with the entire network.
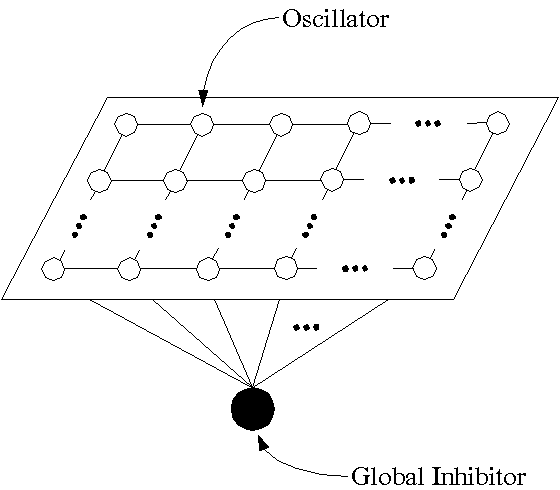
FIGURE 3. Phase plane diagrams illustrating two states of a single oscillator. (a) An oscillatory state occurs when LK of the cubic is above the left part of the sigmoid. (b) A non-oscillatory state occurs when LK is below the left part of the sigmoid. Two fixed points are created on two sides of LK, where the fixed point to the left of LK is stable and acts as a gate to prevent the oscillator from oscillating.
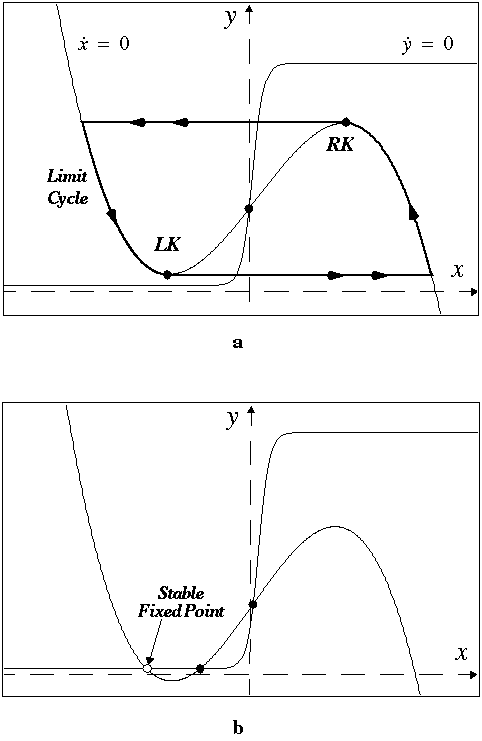
FIGURE 4. Computer simulation of a 20x20 LEGION
network to segment a binary image. (a) The input image with four objects
(arrow, square, cross, and rhombus) and some background noise. (b) Temporal
activity of stimulated oscillators shown for 13,750 integration steps.
Except for the global inhibitor, the vertical axis shows the x value
and the horizontal axis indicates time. The activities of all oscillators
in a group are shown together in one trace. Both synchronization and desynchronization
occur after two cycles, denoted by the dashed line. The networks parameters
are: ![]() ,
, ![]() ,
,
![]() ,
, ![]() ,
,
![]() ,
, ![]() ,
,
![]() ,
,
![]() ,
,
![]() ,
,
![]() ,
, ![]() ,
,
![]() ,
, ![]() ,
,
![]() ,
and
,
and ![]() .
Dynamic weights to a stimulated oscillator are normalized to 8.0.
.
Dynamic weights to a stimulated oscillator are normalized to 8.0.
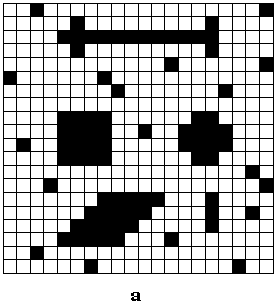
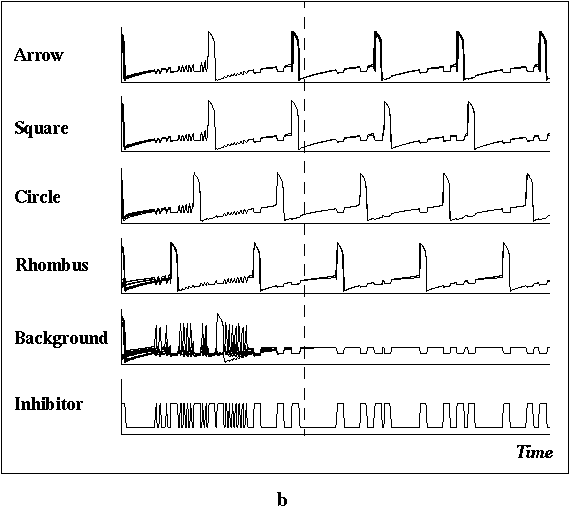
FIGURE 5. The G-LEGION algorithm applied to a 256x256 CT image. (a) Original grey level image showing a horizontal section of a human head. (b) A color map showing the result of segmentation.
a 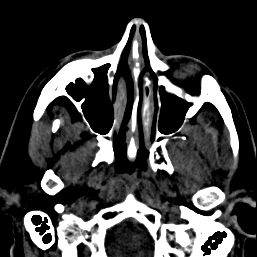 b
b 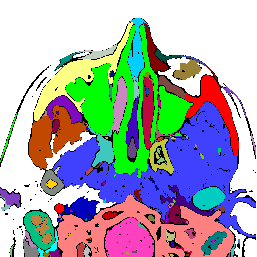
FIGURE 6. G-LEGION applied to a 256x256 T1-weighted MRI image. (a) Original grey level image showing a mid-sagittal section of a human head. (b) A color map showing the result of segmentation.
a 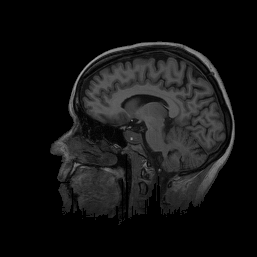 b
b 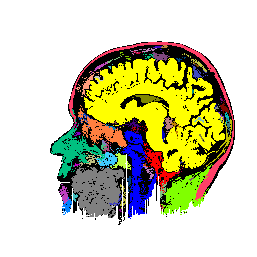
FIGURE 7. Effects of parameters in segmenting
the MRI image of Fig. 6a. The parameters vary
with respect to those used in Fig. 6b. (a) and
(b) show segmentation results when the coupling neighborhood ![]() changes to 8- and 4-neighborhood, respectively. (c) and (d) show segmentation
results when
changes to 8- and 4-neighborhood, respectively. (c) and (d) show segmentation
results when ![]() changes to 24- and 4-neighborhood, respectively. (e) and (f) show segmentation
results when
changes to 24- and 4-neighborhood, respectively. (e) and (f) show segmentation
results when ![]() changes to 7.5 and 2.5, respectively.
changes to 7.5 and 2.5, respectively.
a 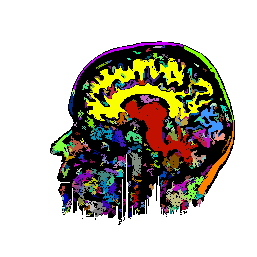 b
b 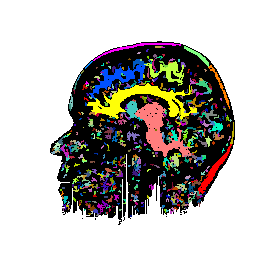
c 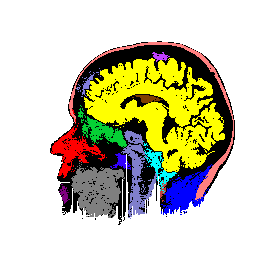 d
d 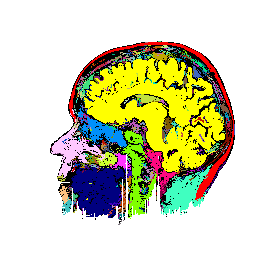
e 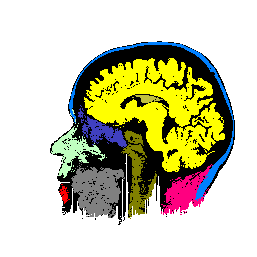 f
f 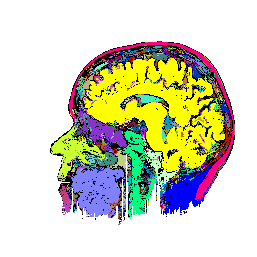
FIGURE 8. G-LEGION applied to a 256x256 T1-weighted MRI image. (a) Original grey level image showing a horizontal section of a human head. (b), (c), and (d) show segmentation results when the tolerance function is cubic, square, and linear, respectively.
a 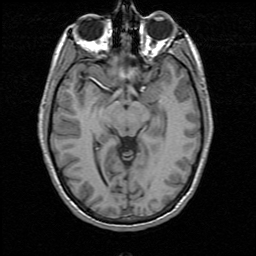 b
b 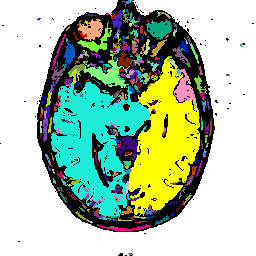
c 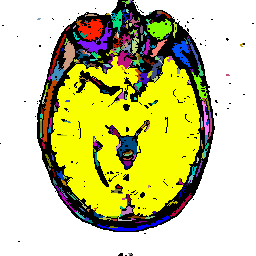 d
d 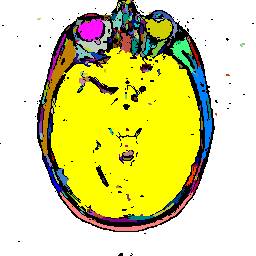
FIGURE 9. Effects of tolerance parameters
in segmenting the MRI image of Fig. 8a. A cubic
tolerance function is used with ![]() clamped to 1. (a), (b), and (c) show segmentation results when
clamped to 1. (a), (b), and (c) show segmentation results when ![]() varies among 80, 40, and 20, respectively.
varies among 80, 40, and 20, respectively.
a 
b 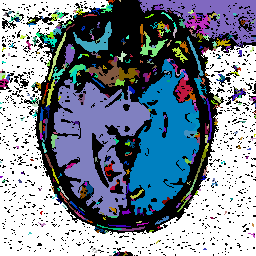 c
c 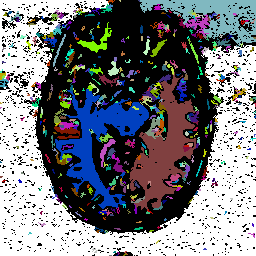
FIGURE 10. Segmentation of a reduced resolution image of Fig. 8a. (a) Original grey level image after a reduction in resolution by half. (b) The result of segmentation.
a 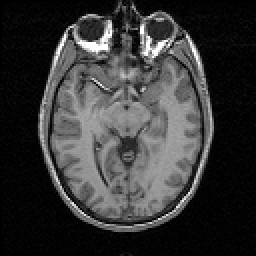 b
b
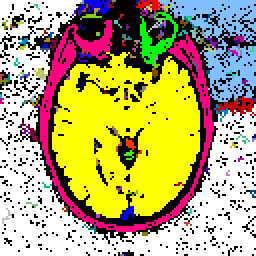
FIGURE 11. G-LEGION applied to 256x256 T1-weighted
MRI images. For all segmentation results a cubic tolerance function is
used. (a) Original grey level image showing a coronal section of a human
head. (b) Segmentation result for the image in (a) with a 24-neighborhood
![]() ,
a 4-neighborhood
,
a 4-neighborhood ![]() ,
,
![]() =
11.5,
=
11.5, ![]() = 1 and
= 1 and ![]() = 316. (c) Original grey level image showing another coronal section of
a human head. (d) Segmentation result for the image in (c) with an 8-neighborhood
= 316. (c) Original grey level image showing another coronal section of
a human head. (d) Segmentation result for the image in (c) with an 8-neighborhood
![]() ,
an 8-neighborhood
,
an 8-neighborhood ![]() ,
,
![]() =
7.5,
=
7.5, ![]() = 7 and
= 7 and ![]() = 90. (e) Original grey level image showing a sagittal section of a human
head. (f) Segmentation result for the image in (e) with a 24-neighborhood
= 90. (e) Original grey level image showing a sagittal section of a human
head. (f) Segmentation result for the image in (e) with a 24-neighborhood
![]() , an
8-neighborhood
, an
8-neighborhood ![]() ,
,
![]() = 17.5,
= 17.5,
![]() =
7 and
=
7 and ![]() = 75. (g) Original grey level image showing another sagittal section of
a human head. (h) Segmentation result for the image in (g) with a 24-neighborhood
= 75. (g) Original grey level image showing another sagittal section of
a human head. (h) Segmentation result for the image in (g) with a 24-neighborhood
![]() ,
an 8-neighborhood
,
an 8-neighborhood ![]() ,
,
![]() =
19.5,
=
19.5, ![]() = 4 and
= 4 and ![]() = 250.
= 250.
a 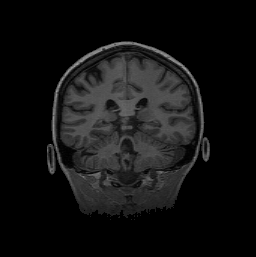 b
b

c 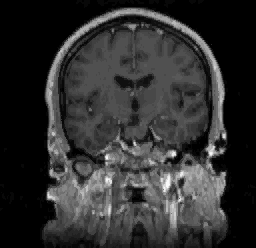 d
d 
e 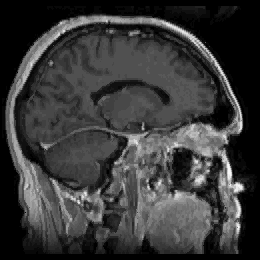 f
f 
g 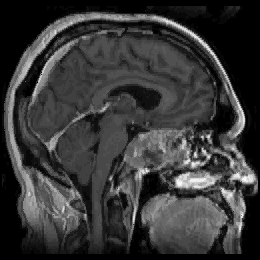 h
h 
FIGURE 12. Segmentation of a 3D MRI volume dataset for extracting the brain. The segmentation results are displayed using volume rendering. (a) and (c) show the results of G-LEGION with a top view (the front facing downward) and a side view (the front facing leftward), respectively. (b) and (d) show the corresponding results produced by slice-by-slice manual segmentation.
a 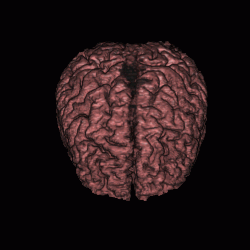 b
b 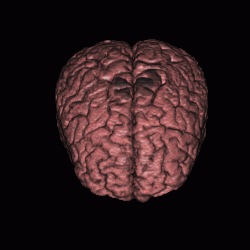
c 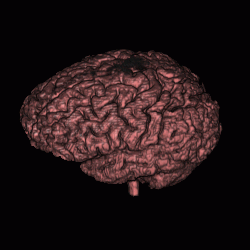 d
d 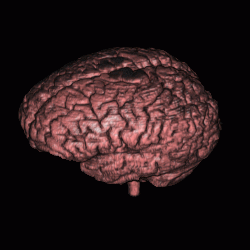
FIGURE 13. Comparison between G-LEGION and manual segmentation. (a) Original grey level image showing a horizontal section of the volume dataset used in Fig. 12. (b) Segmentation result of the image in (a) from 3D segmentation in Fig. 12 by G-LEGION. (c) Result of (b) after tiny holes merge with the brain segment. (d) Segmentation result of the image in (a) from 3D manual segmentation in Fig. 12.
a 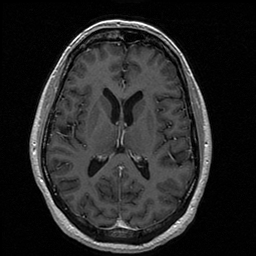 b
b 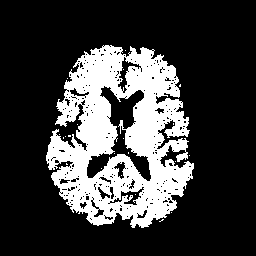
c  d
d 